Piecing together the puzzle: Understanding protein structure can lead to better drug design
Approximately 34% of all drugs approved by the Food and Drug Administration target members of the G protein-coupled receptor family. However, none have been developed to target a specific family known as adhesion G protein-coupled receptors, partially due to how little is known about them. A new study from the laboratory of Demet Araç, PhD, assistant professor in the Department of Biochemistry and Molecular Biology at the University of Chicago, aims to change this by developing a better understanding of how these proteins work.
Adhesion G protein-coupled receptors (aGPCRs) are a wide family of proteins important in neurobiology, development and the immune system in humans and other animals. These proteins work to help cells communicate with each other to make decisions about cell function. “The cells adhere and communicate with one another, and based on this information they migrate, adhere or change how they are developing,” Araç said. “When problems occur with cellular communication events, this can lead to many cancers and neurodevelopmental disorders, such as autism and schizophrenia.” Though scientists have known this for years, because these proteins are so large in size, they have been hard to study despite their importance in so many processes.
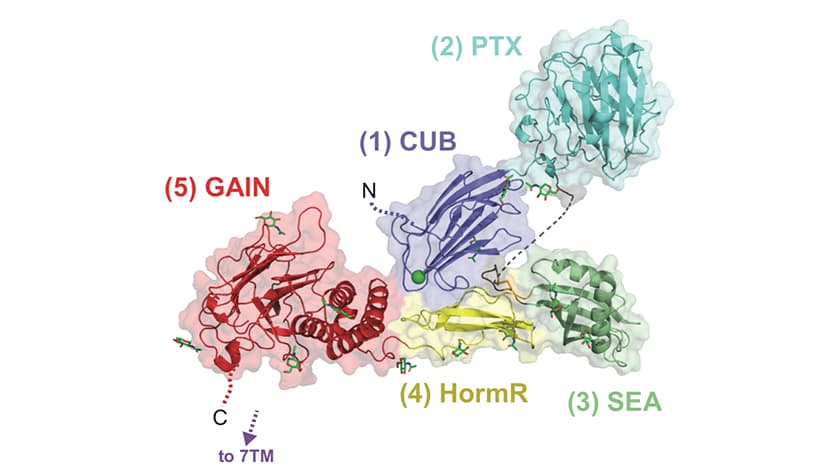
Research by Katherine Leon, PhD, in the Araç laboratory, sheds light on the structure of a member of the aGPCR family — Gpr126 — involved in myelination in the nervous system, ear development and heart development, though exactly how is still unclear. Additionally, mutants of Gpr126 have been linked to cancers and scoliosis. By figuring out the atomic level structure and showing where every individual atom is in 3D space, it’s possible to start answering some of these questions.
The study, published in Nature Communications, used multiple methods, including x-ray crystallography and electron microscopy, to solve the previously unknown structure of the extracellular region (a region of the protein that is outside the cell membrane and in the “free space” between cells) of Gpr126. Leon and colleagues found that this protein could take on multiple shapes, existing in either a closed or open position; bind to calcium ions, which are important in many neuronal pathways; and exist in two forms based on how it was alternatively spliced (which portions of the messenger RNA the cell chose to keep and make into protein.) Based on which spliced version or shape the protein was in, this changed how it was able to signal back to the cell to change functions, the researchers found.
To look at how a specific site of this protein that binds to calcium could control different functions, Leon and collaborators were able to make a mutation in developing zebrafish that makes it so the protein cannot bind calcium. This caused the zebrafish to have defects in ear development and myelination, but not in heart development, showing that how this protein is structured can control different outcomes.
While exactly how Gpr126 is controlling these different processes is still not fully known, these findings show that Gpr126 and other aGPCRs may be promising drug targets for a multitude of different diseases.
“Now that we know, for Gpr126, that these open and closed conformations have different functions, researchers can start figuring out more about these functions differently affect development and disease,” Araç said. “If we know the structure and different conformations of these proteins, scientists can start to design specific drugs to ‘lock’ the protein into one conformation or another to help treat a specific disease, rather than trying randomly until something works. This greatly improves the drug development field.”
The study “Structural basis for adhesion G protein-coupled receptor Gpr126 function” was published on January 10, 2020 in Nature Communications. Additional authors include Joshua A. Riback, Ezra Feldman, Jingxian Li, Tobin R. Sosnick, and Minglei Zhao from The University of Chicago, Rebecca L. Cunningham from Washington University in St. Louis, and Kelly R. Monk from Washington University in St. Louis and currently Oregon Health and Science University.
